Note
Click here to download the full example code
Deconvoluting spectra with pquad¶
This example shows how to deconvolute spectra using the model
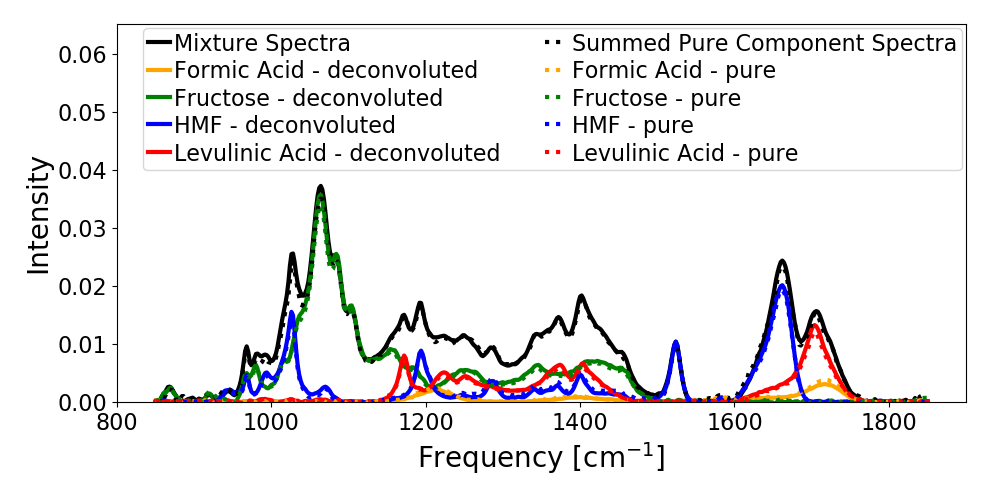
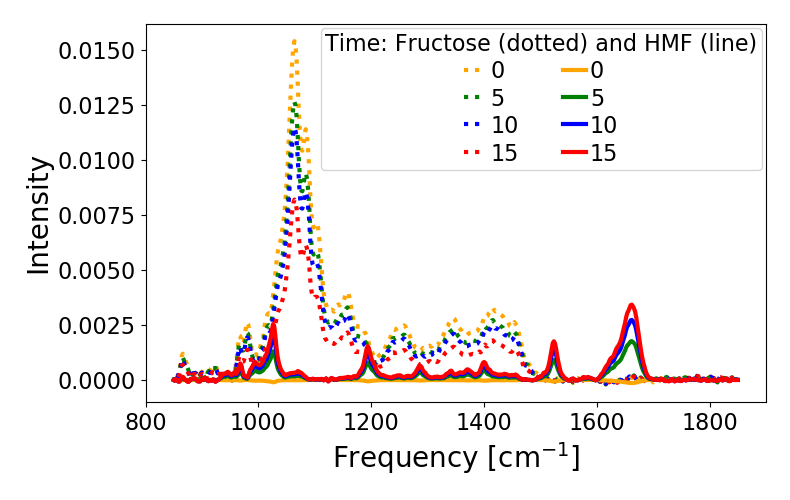
The parity plot for the mixtures where concentrations are known is shown in figure 1 and the plot of concentration with time for the experimental spectra from reacting systems are shown in figure 2 and 3 for different starting concentrations
from __future__ import absolute_import, division, print_function
import os
import numpy as np
import matplotlib.pyplot as plt
from pquad import IR_Results
from pquad import get_defaults
from pquad.plotting_tools import set_figure_settings
Loading data¶
First, we’ll get the default data and load it into pquad. It automatically fits the model to the data in pure_data_path
frequency_range, pure_data_path, mixture_data_path, reaction_data_path = get_defaults()
deconv = IR_Results(4, frequency_range, pure_data_path)
mixture_data_path_file = os.path.join(mixture_data_path, os.listdir(mixture_data_path)[4])
deconv.set_mixture_data(mixture_data_path_file)
deconv_reaction = IR_Results(4, frequency_range, pure_data_path)
deconv_reaction.set_mixture_data(reaction_data_path, contains_concentrations=False)
Set figure settings¶
figure_folder='fit'
set_figure_settings('presentation')
Plot deconvolution for mixture with known species-concentrations¶
Plot comparison of deconvoluted spectra and pure-spectra
deconv.plot_deconvoluted_spectra(figure_folder)
Out:
C:\Users\lansf\Box Sync\Synced_Files\Coding\Python\Github\pQUAD\pquad\pquad.py:802: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
Plot deconvolution of Fructose and HMF during reaction¶
deconvouted_spectra = deconv_reaction.get_deconvoluted_spectra(deconv_reaction.MIXTURE_STANDARDIZED)
for i in range(deconv_reaction.NUM_TARGETS):
if deconv_reaction.PURE_NAMES[i] == 'Fructose':
fructose_index = i
elif deconv_reaction.PURE_NAMES[i] == 'HMF':
HMF_index = i
plt.figure(figsize=(7.9,5))
Colors = ['orange','g','b','r']
for count, spectra in enumerate(np.array(deconvouted_spectra[0])[:,fructose_index][0::5]):
plt.plot(deconv_reaction.FREQUENCY_RANGE,spectra, color=Colors[count], linestyle=':')
for count, spectra in enumerate(np.array(deconvouted_spectra[0])[:,HMF_index][0::5]):
plt.plot(deconv_reaction.FREQUENCY_RANGE,spectra,color=Colors[count], linestyle='-')
plt.legend([str(i) for i in np.arange(len(np.array(deconvouted_spectra[0])[:,fructose_index]))[0::5]] +\
[str(i) for i in np.arange(len(np.array(deconvouted_spectra[0])[:,HMF_index]))[0::5]],title='Time: Fructose (dotted) and HMF (line)',ncol=2)
plt.xlabel('Frequency [cm$^{-1}$]')
plt.ylabel('Intensity')
plt.show()
Out:
C:\Users\lansf\Box Sync\Synced_Files\Coding\Python\Github\pQUAD\examples\deconvolution\plot_deconvolution.py:64: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
Total running time of the script: ( 0 minutes 1.322 seconds)